UNITED STATES
SECURITIES AND EXCHANGE COMMISSION
Washington, D.C. 20549
Form
CURRENT REPORT
Pursuant to Section 13 or 15(d)
of the Securities Exchange Act of 1934
Date of Report (Date of earliest event reported):
(Exact name of registrant as specified in its charter)
| (State or other jurisdiction of incorporation) |
(Commission File Number) |
(IRS Employer Identification No.) |
| (Address of principal executive offices) | (Zip Code) |
Registrant’s telephone number, including area code: +
Check the appropriate box below if the Form 8-K filing is intended to simultaneously satisfy the filing obligation of the registrant under any of the following provisions:
| Written communications pursuant to Rule 425 under the Securities Act (17 CFR 230.425) |
| Soliciting material pursuant to Rule 14a-12 under the Exchange Act (17 CFR 240.14a-12) |
| Pre-commencement communications pursuant to Rule 14d-2(b) under the Exchange Act (17 CFR 240.14d-2(b)) |
| Pre-commencement communications pursuant to Rule 13e-4(c) under the Exchange Act (17 CFR 240.13e-4(c)) |
Securities registered pursuant to Section 12(b) of the Act:
| Title of each class |
Trading symbol |
Name of each exchange on which registered | ||
Indicate by check mark whether the registrant is an emerging growth company as defined in Rule 405 of the Securities Act of 1933 (§230.405 of this chapter) or Rule 12b-2 of the Securities Exchange Act of 1934 (§240.12b-2 of this chapter).
Emerging growth company
If an emerging growth company, indicate by check mark if the registrant has elected not to use the extended transition period for complying with any new or revised financial accounting standards provided pursuant to Section 13(a) of the Exchange Act. ☐
| Item 7.01 | Regulation FD Disclosure. |
From time to time, Wave Life Sciences Ltd. (the “Company”) presents and/or distributes slides and presentations to the investment community to provide updates and summaries of its business. On November 15, 2022, the Company updated its corporate presentation, which is available on the “For Investors & Media” section of the Company’s website at http://ir.wavelifesciences.com/. This presentation is also furnished as Exhibit 99.1 to this Current Report on Form 8-K.
The information in this Item 7.01 is being furnished and shall not be deemed “filed” for purposes of Section 18 of the Securities Exchange Act of 1934, as amended (the “Exchange Act”), or otherwise subject to the liabilities of that Section, nor shall it be deemed incorporated by reference into any registration statement or other filing under the Securities Act of 1933, as amended, or the Exchange Act, except as shall be expressly set forth by specific reference in such filing.
| Item 9.01 | Financial Statements and Exhibits. |
(d) Exhibits
The following exhibit relating to Item 7.01 is furnished and not filed:
| Exhibit No. |
Description | |
| 99.1 | Corporate Presentation of Wave Life Sciences Ltd. dated November 15, 2022 | |
| 104 | Cover Page Interactive Data File (embedded within the Inline XBRL document) | |
SIGNATURES
Pursuant to the requirements of the Securities Exchange Act of 1934, the registrant has duly caused this report to be signed on its behalf by the undersigned hereunto duly authorized.
| WAVE LIFE SCIENCES LTD. | ||
| By: | /s/ Paul B. Bolno, M.D. | |
| Paul B. Bolno, M.D. | ||
| President and Chief Executive Officer | ||
Date: November 15, 2022

Exhibit 99.1 Wave Life Sciences Corporate Presentation November 15, 2022

Forward-looking statements This document contains forward-looking statements. All statements other than statements of historical facts contained in this document, including statements regarding possible or assumed future results of operations, preclinical and clinical studies, business strategies, research and development plans, collaborations and partnerships, regulatory activities and timing thereof, competitive position, potential growth opportunities, use of proceeds and the effects of competition are forward-looking statements. These statements involve known and unknown risks, uncertainties and other important factors that may cause the actual results, performance or achievements of Wave Life Sciences Ltd. (the “Company”) to be materially different from any future results, performance or achievements expressed or implied by the forward-looking statements. In some cases, you can identify forward-looking statements by terms such as “may,” “will,” “should,” “expect,” “plan,” “aim,” “anticipate,” “could,” “intend,” “target,” “project,” “contemplate,” “believe,” “estimate,” “predict,” “potential” or “continue” or the negative of these terms or other similar expressions. The forward-looking statements in this presentation are only predictions. The Company has based these forward-looking statements largely on its current expectations and projections about future events and financial trends that it believes may affect the Company’s business, financial condition and results of operations. These forward-looking statements speak only as of the date of this presentation and are subject to a number of risks, uncertainties and assumptions, including those listed under Risk Factors in the Company’s Form 10-K and other filings with the SEC, some of which cannot be predicted or quantified and some of which are beyond the Company’s control. The events and circumstances reflected in the Company’s forward-looking statements may not be achieved or occur, and actual results could differ materially from those projected in the forward-looking statements. Moreover, the Company operates in a dynamic industry and economy. New risk factors and uncertainties may emerge from time to time, and it is not possible for management to predict all risk factors and uncertainties that the Company may face. Except as required by applicable law, the Company does not plan to publicly update or revise any forward-looking statements contained herein, whether as a result of any new information, future events, changed circumstances or otherwise. 2

UNLOCKING THE BODY’S OWN ABILITY TO TREAT GENETIC DISEASE realizing a brighter future for patients and families 3

Building a leading genetic medicines company LEVERAGING THE ONGOING TARGETING THE TRANSCRIPTOME TO UNLOCK THE GENETIC REVOLUTION BODY’S OWN ABILITY TO TREAT GENETIC DISEASE >6,000 monogenic Increase in Innovative Platform Diversified Pipeline diseases; genetic vastly more Stereopure oligonucleotides CNS: ALS, FTD, HD testing polygenic diseases Novel backbone modifications Muscle: DMD (PN chemistry) Hepatic diseases: AATD Silencing, splicing, and editing modalities Biomarkers to Greater assess target understanding Strong and broad IP engagement of genetic 1 position early in disease and clinical cellular development biology Clinical Expertise GMP Manufacturing Multiple global clinical trials Internal manufacturing capable of producing Many Innovative trial designs Innovations for oligonucleotides at scale diseases precise modification out of reach of transcriptome, for proteome and traditional interactome medicines 4 ALS: Amyotrophic lateral sclerosis; FTD: Frontotemporal dementia; HD: Huntington’s disease; DMD: Duchenne muscular dystrophy; AATD: Alpha-1 antitrypsin deficiency 1 stereopure oligonucleotides and novel backbone chemistry modifications

Wave’s ability to rationally design oligonucleotides enables access to unique disease targets PRISM backbone linkages PO PS PN (Sp) (Rp) - - O S N … … … … … … Phosphoryl guanidine Chirality Chirality Chirality x-ray structure None PN backbone Rp PS backbone Rp PN backbone Sp PS backbone Sp Negative Negative Neutral example charge charge charge PO: phosphodiester PS: phosphorothioate 5
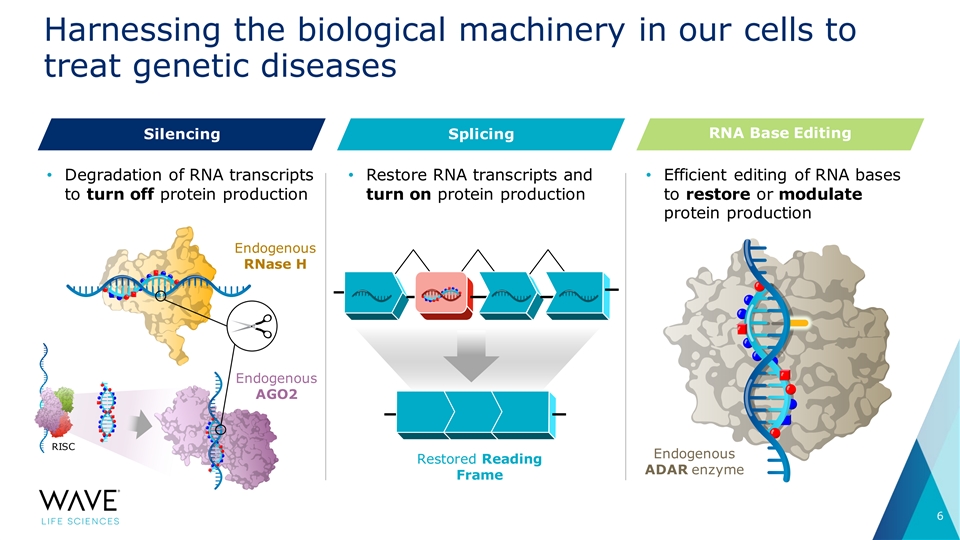
Harnessing the biological machinery in our cells to treat genetic diseases RNA Base Editing Silencing Splicing • Degradation of RNA transcripts • Restore RNA transcripts and • Efficient editing of RNA bases to turn off protein production turn on protein production to restore or modulate protein production Endogenous RNase H Endogenous AGO2 RISC Endogenous Restored Reading ADAR enzyme Frame 6

Unlocking the body’s own ability to treat genetic disease DESIGN OPTIMIZE Chemistry Unique ability to construct Provides the resolution to Sequence stereopure oligonucleotides observe this structural and control three structural interplay and understand how features to efficiently engage it impacts key pharmacological biological machinery properties Stereochemistry Built-for-Purpose Candidates to Optimally Address Disease Biology Silencing | Splicing | RNA Editing 7
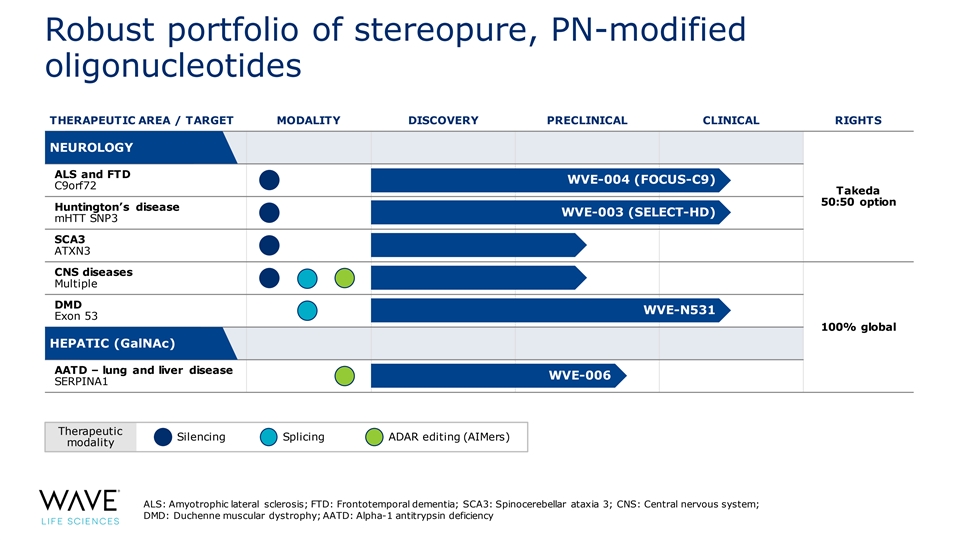
Robust portfolio of stereopure, PN-modified oligonucleotides THERAPEUTIC AREA / TARGET MODALITY DISCOVERY PRECLINICAL CLINICAL RIGHTS N N EE U U R R OL OL OG OG YY ALS and FTD WVE-004 (FOCUS-C9) C9orf72 Takeda 50:50 option Huntington’s disease WVE-003 (SELECT-HD) mHTT SNP3 SCA3 ATXN3 CNS diseases Multiple DMD WVE-N531 Exon 53 100% global H H EEP PA A TT II CC (GalNAc) AATD – lung and liver disease WVE-006 SERPINA1 Therapeutic Silencing Splicing ADAR editing (AIMers) modality ALS: Amyotrophic lateral sclerosis; FTD: Frontotemporal dementia; SCA3: Spinocerebellar ataxia 3; CNS: Central nervous system; DMD: Duchenne muscular dystrophy; AATD: Alpha-1 antitrypsin deficiency 8

WVE-004 Amyotrophic Lateral Sclerosis (ALS) Frontotemporal Dementia (FTD)

C9orf72 repeat expansions: One of the most common genetic causes of ALS and FTD Hexanucleotide (G C )- repeat 4 2 expansions in C9orf72 gene are common autosomal dominate cause for ALS and FTD Typically 100’s- 1000’s of GGGGCC repeats Different manifestations across a clinical spectrum Amyotrophic Lateral Sclerosis (ALS) Frontotemporal Dementia (FTD) • Fatal neurodegenerative disease • Progressive neuronal degeneration in frontal / temporal cortices • Progressive degeneration of motor neurons in • Personality and behavioral changes, gradual brain and spinal cord impairment of language skills • C9-specific FTD: ~10,000 patients in US • C9-specific ALS: ~2,000 patients in US Including patients with C9-associated ALS, FTD or both Sources: Balendra et al, EMBO Mol Med, 2017; Brown et al, NEJM, 2017, DeJesus-Hernandez et al, Neuron, 2011. Renton et al, Neuron, 2011. Zhu et al, Nature Neuroscience, May 2020, Stevens et al, Neurology 1998 10

WVE-004 addresses each biological aspect of C9orf72- associated ALS and FTD RNA variants Disease drivers C9orf72 protein V1 V2 V3 C9orf72 Poly(GP) biomarker 1 Decrease in Genetic mutation Loss-of- selected as preferred beneficial function protein Reduced DPR biomarker Repeat- Wild-type expression expanded C9orf72 ü Abundant in CNS allele allele Dipeptide repeat proteins (DPRs) ü Most soluble 2 Sense: poly(GA), poly(GR) ü Stable expression Antisense: poly(PR), poly(PA) RAN Transcription translation ü Only DPR derived Sense & Antisense: poly(GP) Gain-of- from both sense & function Pathological RNAs RNA foci antisense RNAs Mis-spliced RNA 3 Sense & Stabilized intron 1 Toxic RNA Antisense RNA Antisense aggregation WVE-004 is Variant-selective Preserves C9orf72 Reduces toxic designed to oligonucleotide, protein expression; does gain-of-function affect multiple lowering V1 & V3 not exacerbate potential drivers of drivers of in preclinical loss-of-function driver of disease (RNA 1 toxicity studies disease foci, DPRs) 11 1 Liu et al., 2022 Mol Ther Nuc Acids doi: 10.1016/j.omtn.2022.04.007

ug of oligo / g of tissue ug of oligo / g of tissue Preclinical studies with WVE-004 demonstrated durable reduction of poly(GP) in spinal cord and cortex 6 months after two doses Preclinical in vivo results: C9orf72 protein Six months unchanged at 6 months Spinal Spinal cord 8 150 1.5 1.5 cord ns 6 100 1.0 1.0 4 >90% knockdown of poly(GP) DPR protein 50 0.5 0.5 2 *** ** * *** 0.0 0 0 0.0 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 PBS WVE-004 PBS WVE-004 Weeks 150 Cortex 1.5 Cortex 1.5 8 ns 100 6 1.0 1.0 4 >80% knockdown of 50 0.5 0.5 p≤0.0001 poly(GP) DPR protein 2 0.0 0.0 0 0 PBS WVE-004 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 PBS WVE-004 Weeks Two doses of WVE-004 WVE-004: WVE-004: PBS Poly(GP) DPR Oligonucleotide concentration 12 Liu et al., 2022 Molecular Therapy Nucleic Acids doi: 10.1016/j.omtn.2022.04.007; 2 x 50 ug (day 0, day 7) dosed ICV; DPRs measured by poly(GP) MSD assay. *: p≤ 0.05 **: P ≤ 0.01, ***: P ≤ 0.001. DPR: Dipeptide repeat protein Relative poly(GP) levels Relative poly(GP) levels (normalized to PBS) (normalized to PBS) Relative fold change Re Re la la titviv e f e f olol d c d hc aha nge ng e Relative fold change C9orf72/HPRT1 C9orf72/HPRT1 C9orf72/HPRT1 C9orf72/HPRT1

WVE-004 clinical data demonstrate successful translation of preclinical approach to clinic Target engagement confirmed in patients PK/PD modeling using preclinical in vivo models predicted pharmacodynamically supports advancing FOCUS-C9 clinical study active starting dose ü Poly(GP) reduction in cortex and spinal cord in transgenic mice with WVE-004 ü Sufficient concentrations of WVE-004 in cortex and spinal cord of NHP for target engagement PK: pharmacokinetic PD: pharmacodynamic; Right: Mixed model for repeated measures used to estimate geometric mean ratio to baseline via least squares mean and to calculate p- values. P-values represented by asterisks are for within-dose group geometric mean ratios. *p≤0.05, **p≤0.01, ***p≤0.001. Poly (GP) assay: Wilson et al., 2022 J Neurol Neurosurg 13 Psychiatry doi:10.1136/jnnp-2021-328710. Data presented at ENCALS Meeting (June 1-3, 2022) and International Congress on Frontotemporal Dementias (Nov. 2 – 5, 2022)

Dosing ongoing in FOCUS-C9 clinical trial with multiple doses of WVE-004 Open-label Single dose Multidose extension (OLE) 60 mg n=4 Dose and frequency to be guided by DSMB Target 30 mg engagement n=10 ü OLE clinical trial observed in 20 mg initiated in 4Q 2022 20 mg single dose n=10 n=10 Quarterly doses cohorts 10 mg 10 mg 10 mg n=10 n=6 n=3 Quarterly doses 4 monthly doses Data from all cohorts in the FOCUS-C9 trial are expected in 1H 2023 14

WVE-003 Huntington’s Disease

mHTT toxic effects lead to neurodegeneration, loss of wtHTT functions may also contribute to HD Huntington’s disease (HD) Healthy individual • Wild-type HTT (wtHTT) is critical * for normal neuronal function • Expanded CAG triplet repeat in HTT gene results in production of mutant huntingtin protein (mHTT) wtHTT Stresses • HD is a monogenic autosomal dominant genetic disease; fully Huntington’s disease penetrant and affects entire brain • Fatal disease characterized by cognitive decline, psychiatric illness, and chorea ~50% decrease wtHTT Stresses mHTT + • 30,000 people with HD in the US in wtHTT and more than 200,000 at risk of Loss of wtHTT functions developing HD Synaptic dysfunction | Healthy CNS function Cell death | Neurodegeneration 16

WVE-003: Only investigational HD therapy in clinical development designed to lower mHTT while sparing wtHTT wtHTT supports healthy brain function, WVE-003 especially in the context of stress Regulates synaptic plasticity Unique and innovative wildtype HTT- sparing oligonucleotide Supports synaptic protein Delivered to CNS without invasive transport surgical procedures No complex delivery vehicles required Promotes neuronal survival (e.g. AAV) Designed with next-generation PN Supports cilia and CSF chemistry circulation mHTT, mutant HTT; wtHTT, wild-type HTT; PO, phosphodiester; PS, phosphorothioate; PN, phosphoryl guanidine; wtHTT literature sources: 1. Leavitt 2006 2. Cattaneo 2005 3. Kumar 17 2016 4. Franco-Iborra 2020 5. Hamilton 2015 6. Ochaba 2014 7. Wong 2014 8. Rui 2015 9. Caviston 2007 10. Twelvetrees 2010 11. Strehlow 2007 12. Milnerwood 2010 13. Smith- Dijak 2019 14. Tousley 2019 15. Zhang 2018 16. McAdam 2020 17. Altar 1997 18. Zuccato 2001 19. Gauthier 2004 20. Ferrer 2000 21. Baquet 2004 22. Liu 2011 23. Karam 2015

Allele-selective molecule decreases mHTT, spares wtHTT; Pan-silencer uniformly decreases both Allele-selective activity in CNS of Hu97/18 mice mHTT wtHTT Cortex Striatum 150 150 **** * *** **** *** **** **** **** 100 100 50 50 0 0 4 8 12 4 8 12 4 8 12 4 8 12 4 8 12 4 8 12 Time (weeks) Time (weeks) Pan-silencing Allele-selective PBS Pan-silencing Allele-selective PBS control molecule control molecule Hu97/18 mice administered 3x100 µg intracerebroventricular doses PBS or oligonucleotide. Relative mHTT RNA in cortex (left) striatum (middle) or hippocampus (right) at 4, 8 and 12-weeks post-dosing. Data are mean ± SD, n=8. Stats: ns non-significant, *P<0.05, **P<0.01, ***P<0.0001, 18 ****P<0.0001 versus PBS by 1-way ANOVA. mHTT, mutant HTT; wtHTT, wild-type HTT; Tubb, tubulin Hu97/18 mouse Percentage HTT RNA expression (mHTT/Tubb - PBS)
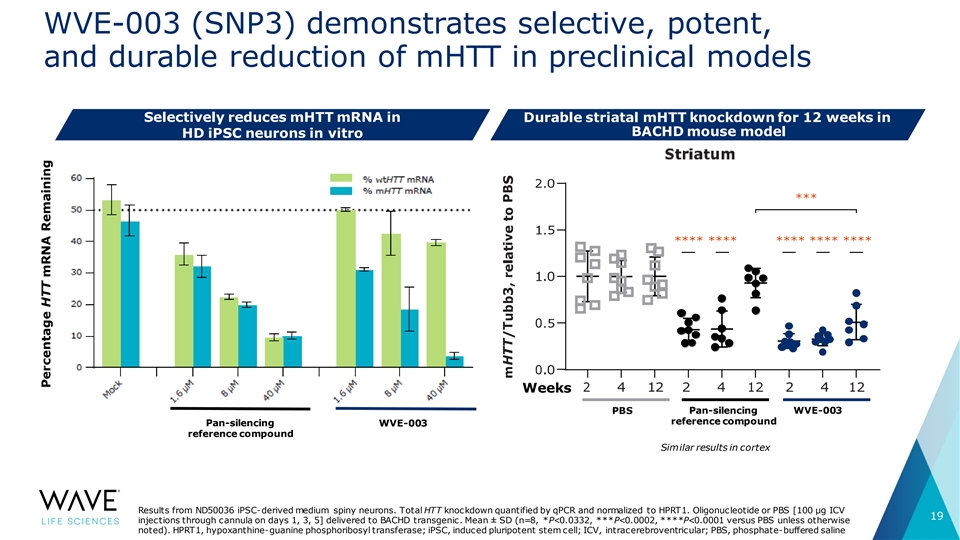
WVE-003 (SNP3) demonstrates selective, potent, and durable reduction of mHTT in preclinical models Selectively reduces mHTT mRNA in Durable striatal mHTT knockdown for 12 weeks in BACHD mouse model HD iPSC neurons in vitro *** **** **** **** **** **** Weeks PBS Pan-silencing WVE-003 reference compound Pan-silencing WVE-003 reference compound Similar results in cortex Results from ND50036 iPSC-derived medium spiny neurons. Total HTT knockdown quantified by qPCR and normalized to HPRT1. Oligonucleotide or PBS [100 μg ICV 19 injections through cannula on days 1, 3, 5] delivered to BACHD transgenic. Mean ± SD (n=8, *P<0.0332, ***P<0.0002, ****P<0.0001 versus PBS unless otherwise noted). HPRT1, hypoxanthine-guanine phosphoribosyl transferase; iPSC, induced pluripotent stem cell; ICV, intracerebroventricular; PBS, phosphate-buffered saline Percentage HTT mRNA Remaining

Initial clinical results indicating allele-selective target engagement suggests translation of preclinical data Reductions in mean CSF mHTT and preservation of wtHTT observed in PK/PD modeling using preclinical pooled analysis of single dose cohorts in SELECT-HD clinical study in vivo models mHTT protein levels wtHTT protein levels Placebo WVE-003 (30 and 60 mg ü Allele selectivity (Hu97/18 mice) pooled*) ü mHTT reduction in cortex and striatum (transgenic mice) ü Concentrations in NHP brain tissues sufficient for target engagement Single dose Single dose of WVE-003 of WVE-003 or placebo or placebo 20 mHTT: mutant huntingtin protein wtHTT: wild-type huntingtin protein *Pooled considering no apparent dose response between 2 cohorts

Expanding single dose cohorts to optimize dose level based on initial clinical results • mHTT protein reductions • Adapting 90 mg Expanding cohort observed in single dose clinical trial to cohorts optimize dose level 60 mg Adding patients to • wtHTT protein levels appear Expanding cohort each cohort consistent with allele- selectivity 30 mg • Generally safe and well- Expanding cohort tolerated Additional single-dose biomarker and safety data are expected in 1H 2023 21 mHTT: mutant huntingtin wtHTT: wild-type huntingtin

WVE-N531 Duchenne muscular dystrophy

Duchenne muscular dystrophy Duchenne muscular dystrophy • Genetic mutation in dystrophin gene prevents the production of dystrophin protein, a critical component of healthy muscle function. • Dystrophin protein established by FDA as surrogate endpoint reasonably likely to predict benefit in 1 patients for accelerated approval in DMD – Confirmatory studies ongoing – Increasing amount of functional dystrophin expression over minimal amount shown with approved therapies is expected to result in greater benefit for patients • Impacts 1 in every 5,000 newborn boys each year; 20,000 new cases annually worldwide. 1 23 Vyondys: www.fda.gov; viltepso; www.fda.gov; Exondys; www.fda.gov; Amondys: www.fda.gov

PN chemistry improved muscle exposure and survival in preclinical mouse models PN boosted muscle concentrations after Treatment with PN-modified molecules led to 100% single dose, which correlated with survival of dKO mice at time of study termination exon-skipping activity 100 Better tissue exposure PS/PO/PN, 150 mg/kg weekly PS/PO/PN, 75 mg/kg bi-weekly 75 PN PS/PO, 150 mg/kg weekly PBS 50 25 PN 0 0 4 8 12 16 20 24 28 32 36 40 Time (weeks) Note: Untreated, age-matched mdx mice had 100% survival at study termination [not shown] Kandasamy et al., 2022; doi: 10.1093/nar/gkac018 24 Survival probability (%)

PS/PO/PN splicing compound restores muscle and respiratory function to wild-type levels in dKO mice Muscle Function Respiratory Function Minute Volume Tidal Volume Specific Force (EDL) 300 150 0.3 200 100 **** 0.2 **** 50 100 0.1 0 0 0.0 21 28 35 42 49 Wild-type 20 40 60 80 100 120 21 28 35 42 49 Age (days) dKO: PBS Age (days) Stimulation Frequency (Hz) dKO: PS/PO/PN Estimated Peak Inspiratory Flow Estimated Peak Expiratory Flow Eccentric Concentration 120 10 6 Wild-type 100 8 dKO / PBS **** 80 **** 4 dKO (PS/PO/PN 6 oligonucleotide) 60 4 40 2 2 20 0 0 0 21 28 35 42 49 21 28 35 42 49 0 1 2 3 4 5 6 7 8 9 10 Age (days) Age (days) Contraction Left: Mdx/utr-/- mice received weekly subQ 150 mg/kg dose of PS/PO/PN stereopure oligonucleotide (postnatal day 10). Age-matched mdx/utr-/- littermates treated with 25 PBS, wild-type C57BL10 mice not treated. Wild-type, dKO PBS mice: 6 wks old; dKO PS/PO/PN: 28 – 41 wks old; Electrophysiology performed at Oxford University based on Goyenvalle et al., 2010 Mol Therapy; Right: Kandasamy et al., 2022; doi: 10.1093/nar/gkac018 2 Specific Force (Nm ) % Force EC0 TVb (ml) mL/s mL/s Minute vol (ml/min)

WVE-N531: Dystrophin restoration in vitro and enhanced muscle distribution in NHPs Dystrophin protein restoration of up to 71% in vitro Enhanced muscle distribution in NHPs Western Blot normalized to Plasma and tissue concentrations of primary healthy human myoblast lysate WVE-N531 (PS/PO/PN) significantly higher than suvodirsen (1st-gen PS/PO) in multiple NHP studies ü Substantially higher muscle concentrations (including heart and diaphragm) as compared to suvodirsen ü Higher plasma Cmax, AUC and Ctrough 26

Top doses evaluated represent human-equivalent level in the range explored in preclinical dKO mouse dKO mouse model Plasma WVE-N531 concentrations compared to plasma suvodirsen concentrations 1000 10 mg/kg - WVE-N531 6 mg/kg - WVE-N531 3 mg/kg - WVE-N531 100 1 mg/kg - WVE-N531 5 mg/kg - Suvodirsen (first-generation PS/PO) 10 1 ü Treatment with PN-modified molecules led to 100% survival of dKO mice at time of study 0.1 termination 0.01 0.001 0.0001 0 10 20 Time post-dose (Day) Pharmacokinetic data demonstrate unique and distinct pharmacological profile with WVE-N531 as compared to first-generation chemistry 27 WVE-N531 open-label clinical trial (NCT04906460) Higher plasma concentrations Plasma Concentration (μg/mL)

Dosing underway with multiple doses of WVE-N531 Ascending intra-patient single doses 3 additional doses every other week Weeks 0 2 4 10 mg/kg 10 mg/kg 10 mg/kg 10 mg/kg Muscle Initial cohort biopsy* Period between dosing 6 mg/kg (~1 – 2 months) •Boys with DMD 3 mg/kg amenable to Data to include: exon 53 skipping• WVE-N531 muscle 1 mg/kg concentrations • WVE-N531 localization • Exon skipping • Dystrophin protein Dose WVE-N531 Clinical data, including muscle biopsies, expected in 4Q 2022 *Biopsies to be taken at least six weeks after initiation of multidosing 28

WVE-006 Alpha-1 antitrypsin deficiency (AATD)

WVE-006: Designed to correct mutant SERPINA1 transcript to address both liver and lung manifestations of AATD WVE-006 designed to correct WVE-006 ADAR editing approach to address key goals of AATD treatment: Z allele mRNA to enable M-AAT protein to be produced 2) Reduce Z-AAT 1) Restore circulating, 3) Retain M-AAT protein aggregation in A functional wild-type M-AAT physiological regulation liver SERPINA1 Z allele mRNA encodes Z-AAT protein with E342K mutation Z-AAT WVE-006 (GalNAc- conjugated AIMer) I(G) RNA correction replaces M-AAT reaches lungs to M-AAT secretion into mutant Z-AAT protein protect from proteases bloodstream with wild-type M-AAT protein Edited SERPINA1 mRNA enables wild- type M-AAT protein production AAT: Alpha-1 antitrypsin Strnad et al., 2020 N Engl J Med 382:1443-55; Blanco et al., 2017 Int J Chron Obstruct Pulmon Dis 12:561-69; Remih 30 et al., 2021 Curr Opin Pharmacol 59:149-56.

~50% RNA editing expected to increase PI*ZZ patient serum AAT levels to PI*MZ levels, with low risk of disease Serum AAT Protein Levels and Risk of AATD by Genotype 100% M-AAT protein 50 ~2.5-7-fold higher 25 100% Z-AAT protein 0 Null ZZ SZ MZ MM Risk of Very High High Low Very Low No Emphysema Risk of Liver No High Possible Possible No Disease ~50% RNA editing 1. Brode, et al. 2012 CMAJ 184:1365-1371; 2. ATS/ERS. 2003 Am J Respir Crit Care Med 168:818–900. 31 Serum AAT protein (µM)

WVE-006 supports dose-dependent RNA editing in human preclinical model systems iPSC-derived human hepatocytes Efficient SERPINA1 editing in donor-derived (ZZ genotype) primary human hepatocytes with WVE-006 (MZ genotype) 80 100 60 WVE-006 90 40 80 20 70 60 0 0.08 0.31 1.25 5.0 50 Mock treated Concentration (µM) 0.0001 0.001 0.01 0.1 1 Dose Dose (uM) Dose Note: Due to MZ genotype, Y-axis ranges from ~50-100% Refresh media 10 Days: 0 2 4 6 8 Collect RNA Left: MZ-donor derived primary human hepatocytes treated with WVE-006 at indicated concentrations for 48 hours Right: Patient-iPSC derived hepatocytes (ZZ genotype) plated on day 0 and treated on day 2 with WVE-006 at indicated concentrations. Media refreshed every 2 days (days 4, 6, 8). 32 RNA was collected on day 10. In each experiment, RNA editing was quantified by Sanger sequencing (n=2 biological replicates) % M-AAT RNA (Mean, s.e.m) % Editing (mean, sem)

WVE-006 results in circulating AAT protein levels 7-fold above PBS control, well above established 11µM threshold WVE-006 treatment results in serum AAT protein SERPINA1 mRNA editing in liver of levels >11 uM in AATD mouse model (NSG-PiZ mice) AATD mouse model (NSG-PiZ mice) (Week 13) ns 2000 60 PBS 1800 WVE-006 1600 WVE-006 (NO LOADING DOSE) 40 1400 1200 ~7-fold 1000 20 increase 800 600 11μM 0 400 200 0 Week WVE-006 administered subcutaneously (10 mg/kg bi-weekly) in 7-week old NSG-PiZ mice (n=5 per group); Loading dose: 3 x 10 mg/kg at Day 0. Left: Liver biopsies collected 33 at week 13 (one week after last dose) and SERPINA1 editing was quantified by Sanger sequencing; Stats: One-way ANOVA with adjustment for multiple comparisons (Tukey); Right: Total serum AAT protein quantified by ELISA; Stats: Two-Way ANOVA with adjustment for multiple comparisons (Tukey) PBS WVE-006 WVE-006 (NO LOADING DOSE) 0 1 2 3 4 5 6 7 8 9 10 11 12 13 Restored AAT protein Serum AAT protein (ug/ml) (Mean, s.e.m) SERPINA1 editing % Editing (Mean, SEM)

WVE-006 leads to restoration of confirmed, wild- type M-AAT protein in serum Overall percentages of serum AAT protein isoforms in NSG-PiZ mice (Week 13) Z-AAT 100 • Mass spectrometry confirms restoration of circulating 80 healthy M-AAT protein in vivo 60 after WVE-006 treatment Wild-type 40 M-AAT protein • Consistent with RNA editing of mutant transcript 20 0 WVE-006 PBS Z-AAT M-AAT WVE-006 administered in 7-week old NSG-PiZ mice (n=5 per group). Relative proportion of M- vs. Z-AAT protein in serum collected from animals at week 34 13 (one week after last dose) was measured by mass spectrometry % Serum AAT isoform (mean, SEM)

Significant increase in neutrophil elastase inhibition activity indicates restored M-AAT protein is functional Serum neutrophil elastase Increased neutrophil elastase inhibition activity inhibition activity demonstrates ns ✱✱✱✱ functionality of AAT protein 100 • Increases in neutrophil elastase, 80 a proteolytic enzyme, may cause emphysema and damage the 60 >3-fold surrounding lung tissue increase • Main function of AAT protein is to 40 over PBS neutralize/control neutrophil 20 elastase 0 Pre-Dose Week 13 PBS WVE-006 GalNAc-conjugated AIMers administered in 7-week old NSG-PiZ mice (n=5 per group). Serum collected from mice was tested for ability to inhibit fixed concentration of neutrophil elastase in an in vitro reaction. Stats: Two-way ANOVA with adjustment for multiple comparisons (Bonferroni) 35 % Relative Elastase inhibition (mean, SEM)

Early lead (pre-optimization) AATD AIMer reduces aggregation of Z-AAT and inflammation in mouse liver Lobular inflammation (19 weeks) 15 25 **** PBS 5 * Early lead AATD AIMer p<0.0001 20 p=0.03 4 10 15 p<0.01 3 5 10 ** 2 5 1 0 4 8 19 0 0 Weeks following first dose AIMer PBS PBS AIMer Early lead pre-optimization AATD AIMer (SA1-5) administered in huADAR/SERPINA1 mice (8–10 wKs old); lower left: 20x liver images PAS-D stained, 19 weeks; Quantification of PAS-D positive staining, Stats 2-way ANOVA; Right: Quantification lobular inflammation grade (Grade based on # of inflammatory foci in lobules: Grade 0: 0; G1 1-5; G2 6-10; 36 G3 11-15; G4 ≥16) and mean globular diameter (40 largest globules/ animal) with HALO. Stats Wilcox ra-n sk um tests %PAS-D positive area % PAS-D positive area (mean±sem) (mean±sem) Mean diameter (µm) Mean Diameter (µm) Inflammation grade Inflammation grade

AIMer-directed editing is highly specific in mice No bystander editing observed on SERPINA1 transcript RNA editing only detected at PiZ mutation site in SERPINA1 transcript RNA editing across transcriptome (mouse liver) (mouse liver) C 0% SERPINA1 PBS (PiZ mutation site) T 100% C 48.2% AATD AIMer T 51.8% % Editing Editing site (PiZ mutation) Dose 3x10 mg/kg (days 0, 2, 4) SC with AATD AIMer (SA1 – 4). Liver biopsies day 7. RNA-seq to quantify on-target SERPINA1 editing, to quantify off-target editing reads mapped to entire mouse genome; plotted circles represent sites with LOD>3 (N=4), SERPINA1 edit site is indicated 37 Coverage Coverage

WVE-006 is a potential first- and best-in-class candidate for AATD • Correct Z-allele mRNA to replace mutant Z-AAT protein with functional wild- type M-AAT protein – RNA editing levels show potential to support conversion of a patient from ZZ to MZ mRNA expression – M-AAT protein can address lung disease – Reduction of Z-AAT protein enables clearance of protein aggregates in liver • M-AAT protein produced with WVE-006 would remain under physiological regulation • mRNA editing is highly specific • Potentially applicable across AATD patient subpopulations • Convenience of subcutaneous administration 38

Planning for clinical development for WVE-006 underway Phase 1/2 placebo-controlled study to establish dose and evaluate target engagement Single-ascending dose (SAD) cohorts Multiple-ascending dose (MAD) cohorts Final cohort in Pi*ZZ patients • Assess dose & frequency • Effective dose to be selected based on PK, PD, and safety Initial cohorts Pi*ZZ pts healthy volunteers Safety, tolerability, PK, change in relevant biomarkers, including serum AAT CTA submissions for WVE-006 expected in 2023 39

AIMers RNA base editing capability
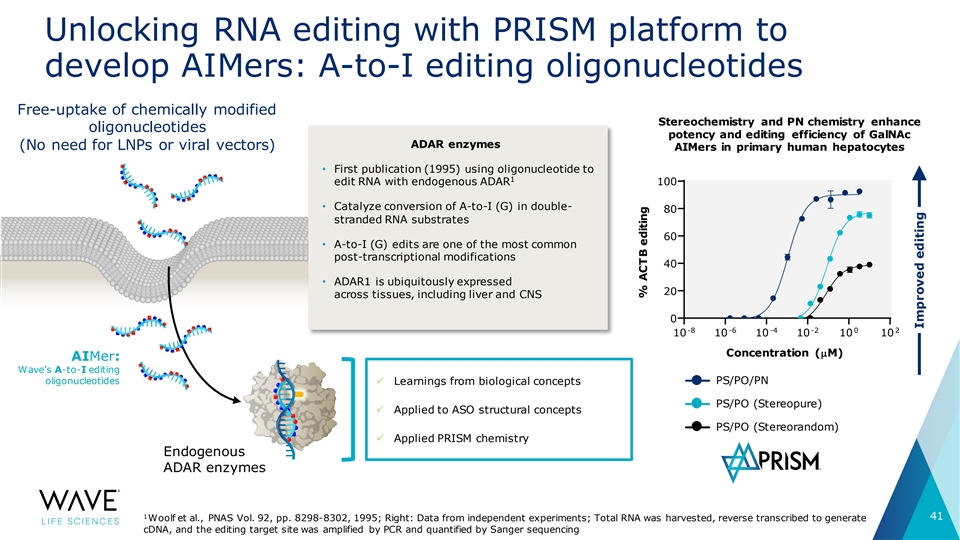
Unlocking RNA editing with PRISM platform to develop AIMers: A-to-I editing oligonucleotides Free-uptake of chemically modified Stereochemistry and PN chemistry enhance oligonucleotides potency and editing efficiency of GalNAc ADAR enzymes (No need for LNPs or viral vectors) AIMers in primary human hepatocytes • First publication (1995) using oligonucleotide to 1 edit RNA with endogenous ADAR 100 • Catalyze conversion of A-to-I (G) in double- 80 stranded RNA substrates 60 • A-to-I (G) edits are one of the most common post-transcriptional modifications 40 • ADAR1 is ubiquitously expressed 20 across tissues, including liver and CNS 0 -8 -6 -4 -2 0 2 10 10 10 10 10 10 Concentration (µM) Concentration (µM) AIMer: Wave’s A-to-I editing oligonucleotides PS/PO/PN ü Learnings from biological concepts PS/PO (Stereopure) ü Applied to ASO structural concepts PS/PO (Stereorandom) ü Applied PRISM chemistry Endogenous ADAR enzymes 1 41 Woolf et al., PNAS Vol. 92, pp. 8298-8302, 1995; Right: Data from independent experiments; Total RNA was harvested, reverse transcribed to generate cDNA, and the editing target site was amplified by PCR and quantified by Sanger sequencing % ACTB editing % Editing Improved editing

AIMers: Realizing potential of therapeutic RNA editing by harnessing endogenous ADAR Solved for key therapeutic attributes for potential best-in-class RNA editing therapeutics Delivery and Efficient ADAR intracellular Stability Beyond liver ü Efficient ADAR recruitment trafficking recruitment • AIMer design • Decade of • GalNAc compatible • AIMer design also principles investment and for targeted liver works for delivery learnings to improve delivery to CNS and other • SAR developed to stability of single- tissue types design AIMers for • Endosomal escape stranded RNAs different targets and nuclear uptake Potent and specific Potential for Subcutaneous IT, IVT, systemic editing in vivo infrequent dosing dosing dosing • Systematized AIMer design enables rapid advancement of new targets • Strong and broad IP in chemical and backbone modifications, stereochemistry patterns, novel and proprietary nucleosides 42 SA R: s tructure-activity relationship

Proof-of-concept preclinical RNA editing data published in Nature Biotechnology (March 2022) • Specificity in vitro & in vivo (NHPs) • GalNAc conjugation • In vitro-in vivo translation (NHPs) • Foundational AIMer SAR AIMers detected in liver of NHP at Day 50 Substantial and durable editing in NHP ADAR editing with ACTB AIMer is (PK) liver in vivo highly specific (PD) RNA editing within full transcriptome (primary human hepatocytes) GalNAc AIMers Day 50 RNA editing ACTB in NHP GalNAc AIMers % Editing 43 RNA editing only detected at editing Monian et al., 2022 published online Mar 7, 2022; doi: 10.1038.s41587-022-01225-1 site in ACTB transcript SAR structure-activity relationship Confidence (LOD score)

Systemic in vivo editing without delivery vehicles Substantial RNA editing across multiple tissues following single subcutaneous dose of UGP2 AIMer Control Specific liver associated cells UGP2 AIMer (unconjugated) T cell Cholan- Macro- NK cell LSEC giocyte phage subset Editing: Potent, durable, specific A à I (G) RNA editing Delivery: Efficient RNA editing in preclinical in vivo models: ü Targeted delivery (GalNAc) ü Systemic delivery ü Local delivery (IT, IVT, others) Editing without GalNAc conjugation Potential to accelerate timelines to candidate with AIMer pipeline expansion 44 Right: Single dose of 100mg/kg unconjugated UGP2 AIMer, seven days post dose; WAT: White adipose tissue; BAT: Brown adipose tissue; CD3+: T-cells and subset of NK cells; EpCAM+(Epithelial cell adhesion molecule): mainly cholangiocytes within liver; LSEC cells (Liver Sinusoidal Endothelial Cells); M0 cells: macrophages

Substantial in vivo editing without delivery vehicles in CNS tissues UGP2 AIMer-1 Peak RNA editing observed one-month post-single dose across tissues PBS Peak 30% >40% 25% >40% 50% >65% editing Potential CNS editing targets to benefit from learnings taken from clinical CNS silencing programs Transgenic huADAR mice administered 100 µg AIMer or PBS on day 0 and evaluated for UGP2 editing across CNS tissues at 1, 4, 8, 12, and 16-weeks post dose. 45 Percentage UGP2 editing determined by Sanger sequencing. Stats: 2-way ANOVA compared to PBS (n=5 per time point per treatment) *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. ICV intracerebroventricular; PBS phosphate buffered saline

Expanding addressable disease target space using AIMers to activate pathways and upregulate expression Correct G-to-A driver mutations with AIMers Modulate protein interactions with AIMers Modulate protein- Restore or correct Achieved protein interaction protein function POC Upregulate expression WVE-006 Modify function (GalNAc AIMer) AATD Post-translational modification Alter folding or processing AIMers provide dexterity, with applications beyond precise correction of genetic mutations, including upregulation of expression, modification of protein function, or alter protein stability 46 POC: proof of concept

Dose dependent modulation of protein/protein interactions Dose-dependent gene upregulation (NQO1) in vitro following Nrf2 editing to disrupt protein/protein interaction KEAP1 RNA editing efficiency Gene upregulation NQO1 qPCR NRF2 % Editing 0.0016uM NRF2 is 0.0016uM 0.008uM degraded by 0.008uM proteasome 0.04uM 0.04uM 0.2uM Transcription is repressed 0.2uM 1uM 1uM 5uM 5uM Basal conditions WV50603 0.0016uM WV50604 0.0016uM 0.008uM 0.008uM 0.04uM KEAP1 0.04uM 0.2uM 0.2uM ADAR editing site 1uM 1uM WV40590 5uM 5uM 0.0016uM 0.0016uM 0.008uM AIMer NRF2 is stabilized 0.008uM 0.04uM 0.04uM Transcription is activated 0.2uM NRF2 0.2uM 1uM 1uM 5uM 5uM ADAR-modified conditions 0 2 4 6 8 0 20 40 60 80 100 NQO1 mRNA Fold Change to non-targeting control % Editing 47 n=2; Primary hepatocytes 48h of treatment with the indicated dose concentration of AIMers UGP2 (Control) NRF2 AIMers AIMer

AIMers enable activation of gene pathway in vivo with single edit NRF2 downstream gene upregulation following GalNAc AIMer Nrf2 liver editing in vivo Nrf2 mRNA editing in vivo in liver of mRNA editing in vivo in liver of mice mice with GalNAc AIMers Nrf2 activation Nrf2 activation of of NQO1 expression GSTM1 expression 80 8 8 *** **** **** **** 6 6 60 **** **** 4 4 ** 40 2 2 * 0 0 20 0 RNAseq transcriptome analysis confirms disruption of Nrf2 protein interaction with upregulation of key factors UGP2 AIMer AIMer 2 Note: Editing percentage for UGP2 control AIMer AIMer 1 indicates editing of UGP2 mRNA Methods: hADAR C57BL/6 mice dosed subQ (days 0, 2, 4) at 10mg/kg GalNAc-conjugated AIMers. Livers harvested (day 7), analyzed for editing and NQO1 expression via Sanger sequencing or qPCR, respectively. Data analyzed via One-way ANOVA with Tukey’s multiple comparison test. Asterisks indicate statistical significance to PBS-treated animals as 48 follows: * = p<0.05; ** = p<0.01; *** = p<0.001; **** = p<0.0001 PBS Nrf2 AIMer 1 Nrf2 AIMer 2 UGP2 AIMer PBS Nrf2 AIMer 1 Nrf2 AIMer 2 UGP2 AIMer PBS Nrf2 AIMer 1 Nrf2 AIMer 2 UGP2 AIMer RNA editing efficiency Percent mRNA editing Gene upregulation Relative Expression (NQO1/HPRT) Relative Expression (GSTM1/HPRT)

Upregulation: AIMers can edit RNA motifs to restore or upregulate gene expression RNA binding proteins recognize sequence motifs to regulate various mRNA properties Stability Transport Processing Protein production • Enhance or inhibit • Intracellular localization • Splicing • Translational efficiency • PolyA usage mRNA decay • Capping AIMer edits mRNA à “dials up” gene expression No binding RNA binding protein I(G) Edited mRNA A mRNA Decay 49

AIMers can edit RNA motifs to upregulate gene expression in hepatocytes and T-cells in vitro Editing RNA Motifs to regulate RNA half-life to upregulate RNA expression is possible for clinically-relevant targets, including both metabolic and immune targets Target A Target B Target C Target D 15 5 8 8 4 6 6 10 3 4 4 AIMer 2 5 2 2 1 0 0 0 0 AIMER-1 AIMER-2 Mock AIMER-1 AIMER-2 Mock AIMER-1 AIMER-2 Mock AIMER-1 AIMER-2 Mock Primary human hepatocytes (in vitro) Primary human T-cells (in vitro) Achieving >2-fold mRNA upregulation in vitro across multiple different targets with AIMer editing 50 Gene upregulation Fold increase Target A mRNA Fold increase Target B mRNA Fold increase Target C mRNA Fold increase Target D mRNA
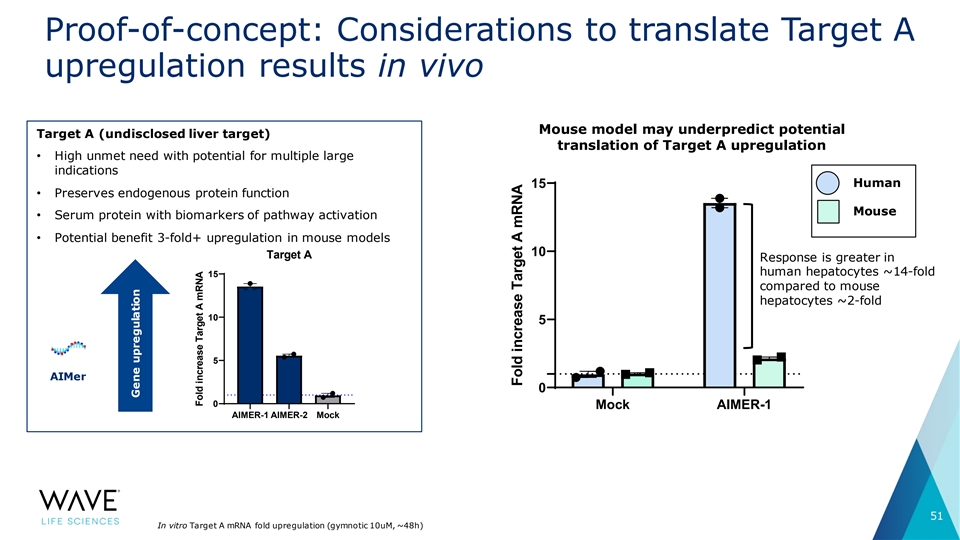
Proof-of-concept: Considerations to translate Target A upregulation results in vivo Mouse model may underpredict potential Target A (undisclosed liver target) translation of Target A upregulation • High unmet need with potential for multiple large indications Human 15 • Preserves endogenous protein function Mouse • Serum protein with biomarkers of pathway activation • Potential benefit 3-fold+ upregulation in mouse models 10 Target A Response is greater in human hepatocytes ~14-fold 15 compared to mouse hepatocytes ~2-fold 10 5 5 AIMer 0 0 Mock AIMER-1 AIMER-1 AIMER-2 Mock 51 In vitro Target A mRNA fold upregulation (gymnotic 10uM, ~48h) Gene upregulation Fold increase Target A mRNA Fold increase Target A mRNA

AIMers upregulate mRNA and downstream serum protein in vivo above anticipated threshold Target A mRNA editing mRNA upregulation Protein upregulation (undisclosed liver 7 days post-initial dose 7 days post-initial dose 7 days post-initial dose target) GalNAc AIMer GalNAc AIMer GalNAc AIMer 20 • High unmet need ** * with potential for 20 * ** 80 multiple large 15 indications 15 60 • Preserves endogenous protein 10 10 function 40 • Serum protein with 5 5 biomarkers of 20 pathway activation 0 • Potential benefit 3- 0 0 PBS AIMER-1 AIMER-2 PBS AIMER-1 AIMER-2 PBS AIMER-1 AIMER-2 fold+ upregulation in mouse Potential threshold for benefit ü In vitro to in vivo translation of mouse Target A mRNA upregulation ü In vivo mRNA upregulation corresponds to an upregulation of Target A protein in serum at Day 7 demonstrating proof-of-concept 52 hADAR mouse dosed subcutaneously 3 x 10 mg/kg GalNAc-conjugated AIMer or PBS days (0, 2, 4), taken down at day 7 RNA editing Percent Editing Upregulation Target A mRNA fold change Fold increase pre-post dose Serum Target A (pg/mL)

RNA editing of nonsense mutation found in MECP2 (Rett Syndrome) restores functional protein Nonsense mutations found in Rett Syndrome can occur in multiple locations on RNA transcript: Normal: … CGA… wild type protein Variant base Rett Syndrome: … TGA… premature stop codon ADAR editing site ADAR editing: … TGG… restored protein in vitro ADAR editing of over 60% targeting Full length MECP2 protein is expressed MECP2 disease transcript following ADAR editing PN chemistry improved editing Dose-dependent RNA editing of efficiency in vitro MECP2 mutation with PS/PN AIMer PS/PN AIMer 100 WV-40573 + Dosed with hADAR hADAR 100 WV-40573 - hADAR Control (no hADAR) 80 ADAR Edited MECP2 80 Endogenous MECP2 60 60 Loading Control 40 40 20 20 0 0 mock PS PS-PN 0.0001 0.001 0.01 0.1 1 10 100 [nM] 293T cells transfected with both nonsense mutation on MECP2 (GFP-fusion construct) and ADAR plasmids. AIMers transfected for 48h prior to RNA extraction and sequencing. Percentage editing determined by Sanger sequencing. Left: Single dose (25nM) treatment Middle: Full dose response curve 53 (25nM, 5-fold dilution, 48h treatment) in presence or absence of hADAR Right: Western blot for MECP2 protein. Three biological replicates, NTC AIMer, mock and naïve 293T cells probed for fusion protein. Percentage A→ G editing Percentage A→ G editing Ladder NTC Mock Naive

Wave’s discovery and drug development platform

Improvements in PRISM primary screen hit rates accelerate drug discovery over time Primary screen hit rates with silencing far above industry standard hit rates Chemistry, PN stereochemistry & machine learning optimization 100 80 80.0% (2020 - current) 60 55.4% (2019) 40 Stereopure 32.9% 20 Stereorandom 12.2% 0 Chemistry improvements and PRISM advancement All screens used iPSC-derived neurons; Data pipeline for improved standardization. Hit rate = % of oligonucleotides with target 55 knockdown greater than 50%. Each screen contains >100 oligonucleotides. ML: machine learning % Hit Rate

Potency is enhanced with addition of PN modifications across modalities Silencing Splicing Editing Target knockdown (% remaining) % Skipping % Editing 100 80 60 40 20 0 -8 -6 -4 -2 0 2 10 10 10 10 10 10 Concentration (µM) Ranked by potency of reference Concentration (µM) PS/PO compound Ranked by potency of reference PS/PO compound PS/PO/PN PS/PO (Stereopure) PS/PO reference compound PS/PN modified compound PS/PO (Stereorandom) Left: Experiment was performed in iPSC-derived neurons in vitro; target mRNA levels were monitored using qPCR against a control gene (HPRT1) using a 56 linear model equivalent of the ∆∆Ct method; Middle: DMD patient-derived myoblasts treated with PS/PO or PS/PO/PN stereopureoligonucleotide under free-uptake conditions. Exon-skipping efficiency evaluated by qPCR. Right: Data from independent experiments Improved knockdown Improved skipping % Editing Improved editing

Adding PN chemistry modifications to C9orf72- targeting oligonucleotides improved potency in vivo Cortex Spinal Cord C9orf72-targeting oligonucleotides PS/PO backbone chemistry PS/PO/PN backbone chemistry Exposure (µg/g) Exposure (µg/g) Improved tissue exposure Oligonucleotide concentrations quantified by hybridization ELISA. Graphs show robust best fit lines with 95% confidence intervals (shading) for PK-PD 57 analysis; Liu et al. Molecular Therapy Nucleic Acids 2022; Kandasamy et al., Nucleic Acids Research, 2022, doi: 10.1093/nar/gkac037 %C9orf72 V3 transcript remaining Improved knockdown

PN chemistry improves distribution to CNS Distribution of oligonucleotides in non-human primate CNS 1-month post single IT dose Cerebral Cortex Striatum Oligonucleotide (red staining) PS/PO Backbone chemistry PS/PO/PN Midbrain Cerebellum Hippocampus Spinal cord 58 NHPs administered 1x12 mg oligonucleotide or PBS by intrathecal injection/lumbar puncture (IT). CNS tissue evaluated 11 or 29 days after injection (n=6 per group). Oligonucleotide was visualized by ViewRNA (red), and nuclei are counterstained with hematoxylin. Images from day 29.

PRISM PN siRNA led to unprecedented silencing >3 months after single dose Ago Ago 2 2 l loo ada indi g ng (liver, transgenic mice) (liver, transgenic mice) 25 HSD17B13 mRNA Wk2 Wk 7 Wk 14 (liver, transgenic mice) 20 125 15 100 75 10 50 * * 5 **** 25 0 0 0 2 4 6 8 10 12 14 16 Time (weeks) Reference 2 PBS HSD PN/PS/PO HSD Reference 2 HSD PN/PS/PO Mice expressing a human HSD17B13 transgene were treated with 3 mg/kg of the indicated siRNA or PBS, and liver mRNA, guide strand concentration, and Ago2 loading were quantified at the indicated times post-dose. Stats: Two-way ANOVA with post-hoc test * P<0.05, ****P<0.0001. Reference 2 is 59 based on Foster, et al., 2018. Mol. Ther. 26, 708-717 % mRNA remaining (HSD17B13/Hprt) Fold change relative to Reference 2

Established internal GMP manufacturing for multiple oligonucleotide modalities Strong technical knowhow Established and operating expertise infrastructure • Experienced team led by Sridhar • State of the art facilities (90,000 Vaddeboina, PhD (SVP sq ft) and expansion space Chemistry, Manufacturing, • Process and analytical Controls) development labs • Experts in oligonucleotide • GMP oligonucleotide (API) synthesis (ASOs, DNAs, RNAs, manufacturing siRNAs) • Established Quality and GMP • Proven track record scaling systems (QA, supply chain, complex chemistries; delivered logistics, QC testing) clinical supply for six programs at Wave Scalable to support Wave’s GMP manufacturing needs, as well as potential new partners 60

Upcoming milestones 61

Steady flow of data updates expected to further inform future opportunities and unlock value WVE-004ü Delivered clinical target engagement data with single doses C9orf72 ALS ü Initiated OLE clinical trial in 4Q 2022 CNS & FTD • Data from all cohorts in FOCUS-C9 trial expected in 1H 2023 Silencing (Intrathecal) ü Delivered single-dose clinical data indicating reduction in mHTT WVE-003 with wtHTT preserved, appearing consistent with allele-selectivity HD SNP3 • Additional single-dose biomarker and safety data in 1H 2023 WVE-N531 • Clinical data, including muscle biopsies, to enable decision Muscle DMD Exon Splicing making in 4Q 2022 (IV) 53 ü Selected an AATD AIMer development candidate and initiated WVE-006 IND-enabling activities AATD Targeted • Submit clinical trial applications in 2023 ADAR delivery liver editing (Subcutaneous) WVE-004 FOCUS-C9 clinical trial (NCT04931862); WVE-003 SELECT-HD clinical trial (NCT05032196); WVE-N531 open-label 62 clinical trial (NCT04906460)

Realizing a brighter future for people affected by genetic diseases For more information: Kate Rausch, Investor Relations InvestorRelations@wavelifesci.com 617.949.4827 63
